Using Apache Spark for Data Processing and Machine Learning to detect nature of physical activity using sensor data.
The PAMAP2 Physical Activity Monitoring dataset contains data of 18 different physical activities, performed by 9 subjects wearing 3 inertial measurement units and a heart rate monitor. This classification task aims to distinguish activities of minimum,low,medium and high effort based on various physical activities sensor measurements given in the dataset.Check the raw data formatting in the below cell for more details.
We will load the data into Hadoop ditributed file system and utilize pyspark to process the data and use mlib library in pyspark for machine learning algorithms to detect the physical activity based on readings.
Raw data format
Synchronized and labeled raw data from all the sensors (3 IMUs and the HR-monitor) is merged
into 1 data file per subject per session (protocol or optional), available as text-files (.dat).
Each of the data-files contains 54 columns per row, the columns contain the following data:
– 1 timestamp (s)
– 2 activityID (see II.2. for the mapping to the activities)
– 3 heart rate (bpm)
– 4-20 IMU hand
– 21-37 IMU chest
– 38-54 IMU ankle
The IMU sensory data contains the following columns:
– 1 temperature (°C)
– 2-4 3D-acceleration data (ms-2), scale: ±16g, resolution: 13-bit
– 5-7 3D-acceleration data (ms-2), scale: ±6g, resolution: 13-bit*
– 8-10 3D-gyroscope data (rad/s)
– 11-13 3D-magnetometer data (μT)
– 14-17 orientation (invalid in this data collection)
- Activity IDs: – 1 lying – 2 sitting – 3 standing – 4 walking – 5 running – 6 cycling – 7 Nordic walking – 9 watching TV – 10 computer work – 11 car driving – 12 ascending stairs – 13 descending stairs – 16 vacuum cleaning – 17 ironing – 18 folding laundry – 19 house cleaning – 20 playing soccer – 24 rope jumping – 0 other (transient activities) Note: data labeled with activityID=0 should be discarded in any kind of analysis. This data mainly covers transient activities between performing different activities, e.g. going from one location to the next activity’s location, or waiting for the preparation of some equipment. Also, different parts of one subject’s recording (in the case when the data collection was aborted for some reason) was put together during these transient activities (noticeable by some “jumping” in the HR-data).
For the classification task we have binned the activity list id’s given above based on the intensity of each physical activitie and this would be used as our target variable for classification:
Minimun: [lying,sitting,standing,computer work,car driving,folding laundry] minimum_activity = [1,2,3,9,10,11,18] Low: [descending stairs, ironing, house cleaning] low_activity = [13,17,19] medium: [walking,descending stairs,vacuum cleaning] medium_activity = [4,12,16] high: [running,cycling,nordic walking,playing soccer,rope jumping] high_activity = [5,6,7,20,24]
Move Rawdata to HDFS and Convert into ORC format
Follow the below steps to download and move raw data from local file system to hdfs and convert into orc format for distributed processing
Download the dataset
[hadoop@test-ser-vm01 pamap2]$ wget http://archive.ics.uci.edu/ml/machine-learning-databases/00231/PAMAP2_Dataset.zip
[hadoop@test-ser-vm01 pamap2]$ unzip PAMAP2_Dataset.zip
[hadoop@test-ser-vm01 pamap2]$ cd PAMAP2_Dataset
[hadoop@test-ser-vm01 pamap2]$ cd PAMAP2_Dataset/Protocol
[hadoop@test-ser-vm01 Protocol]$ ls -lrt
total 1271440
-rw-------. 1 hadoop hadoop 141698539 Jan 10 2012 subject101.dat
-rw-------. 1 hadoop hadoop 207349310 Jan 10 2012 subject102.dat
-rw-------. 1 hadoop hadoop 117863364 Jan 10 2012 subject103.dat
-rw-------. 1 hadoop hadoop 153227482 Jan 10 2012 subject104.dat
-rw-------. 1 hadoop hadoop 173773599 Jan 10 2012 subject105.dat
-rw-------. 1 hadoop hadoop 168263273 Jan 10 2012 subject106.dat
-rw-------. 1 hadoop hadoop 145823268 Jan 10 2012 subject107.dat
-rw-------. 1 hadoop hadoop 190019653 Jan 10 2012 subject108.dat
-rw-------. 1 hadoop hadoop 3891140 Jan 10 2012 subject109.dat
Create directory in HDFS and copy files from local to HDFS
[hadoop@test-ser-vm01 Protocol]$ hadoop fs -mkdir /mnt/data/scripts/notebooks/ashwin/data
[hadoop@test-ser-vm01 Protocol]$ hadoop dfs -put subject* /mnt/data/scripts/notebooks/ashwin/data
[hadoop@test-ser-vm01 Protocol]$ hadoop fs -ls /mnt/data/scripts/notebooks/ashwin/data
Found 9 items
-rw-r--r-- 3 hadoop supergroup 141698539 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject101.dat
-rw-r--r-- 3 hadoop supergroup 207349310 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject102.dat
-rw-r--r-- 3 hadoop supergroup 117863364 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject103.dat
-rw-r--r-- 3 hadoop supergroup 153227482 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject104.dat
-rw-r--r-- 3 hadoop supergroup 173773599 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject105.dat
-rw-r--r-- 3 hadoop supergroup 168263273 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject106.dat
-rw-r--r-- 3 hadoop supergroup 145823268 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject107.dat
-rw-r--r-- 3 hadoop supergroup 190019653 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject108.dat
-rw-r--r-- 3 hadoop supergroup 3891140 2018-03-14 08:41 /mnt/data/scripts/notebooks/ashwin/data/subject109.dat
Create HIVE external Table with schema and Load data
hive> CREATE SCHEMA IF NOT EXISTS test1;
OK.
hive>CREATE EXTERNAL TABLE IF NOT EXISTS test1.pamap2_table
(time DOUBLE, activity DOUBLE, heart_rate DOUBLE,hand_temp DOUBLE,hand_acc16g_1 DOUBLE, hand_acc16g_2 DOUBLE, hand_acc16g_3 DOUBLE, hand_acc6g_1 DOUBLE, hand_acc6g_2 DOUBLE, hand_acc6g_3 DOUBLE, hand_gyro_1 DOUBLE,hand_gyro_2 DOUBLE,hand_gyro_3 DOUBLE, hand_mag_1 DOUBLE, hand_mag_2 DOUBLE, hand_mag_3 DOUBLE, hand_orient_1 DOUBLE, hand_orient_2 DOUBLE, hand_orient_3 DOUBLE, hand_orient_4 DOUBLE,
chest_temp DOUBLE,chest_acc16g_1 DOUBLE, chest_acc16g_2 DOUBLE, chest_acc16g_3 DOUBLE, chest_acc6g_1 DOUBLE, chest_acc6g_2 DOUBLE, chest_acc6g_3 DOUBLE, chest_gyro_1 DOUBLE,chest_gyro_2 DOUBLE,chest_gyro_3 DOUBLE, chest_mag_1 DOUBLE, chest_mag_2 DOUBLE, chest_mag_3 DOUBLE, chest_orient_1 DOUBLE, chest_orient_2 DOUBLE, chest_orient_3 DOUBLE, chest_orient_4 DOUBLE,
ankle_temp DOUBLE,ankle_acc16g_1 DOUBLE, ankle_acc16g_2 DOUBLE, ankle_acc16g_3 DOUBLE, ankle_acc6g_1 DOUBLE, ankle_acc6g_2 DOUBLE, ankle_acc6g_3 DOUBLE, ankle_gyro_1 DOUBLE,ankle_gyro_2 DOUBLE,ankle_gyro_3 DOUBLE, ankle_mag_1 DOUBLE, ankle_mag_2 DOUBLE, ankle_mag_3 DOUBLE, ankle_orient_1 DOUBLE, ankle_orient_2 DOUBLE, ankle_orient_3 DOUBLE, ankle_orient_4 DOUBLE )
ROW FORMAT DELIMITED
FIELDS TERMINATED BY ' '
STORED AS TEXTFILE
LOCATION '/mnt/data/scripts/notebooks/ashwin/data/';
OK.
The import can be verified by listing the first few rows in the table:
hive> select * from test1.pamap2_table limit 2;
OK
8.38 0.0 104.0 30.0 2.37223 8.60074 3.51048 2.43954 8.76165 3.35465 -0.0922174 0.0568115 -0.0158445 14.6806 -69.2128 -5.58905 1.0 0.0 0.0 0.0 31.8125 0.23808 9.80003 -1.68896 0.265304 9.81549 -1.41344 -0.00506495 -0.00678097 -0.00566295 0.47196-51.0499 43.2903 1.0 0.0 0.0 0.0 30.3125 9.65918 -1.65569 -0.0997967 9.64689 -1.55576 0.310404 0.00830026 0.00925038 -0.0175803 -61.1888 -38.9599 -58.1438 1.0 0.0 0.0 0.0
8.39 0.0 NaN 30.0 2.18837 8.5656 3.66179 2.39494 8.55081 3.64207 -0.0244132 0.0477585 0.00647434 14.8991 -69.2224 -5.82311 1.0 0.0 0.0 0.0 31.8125 0.31953 9.61282 -1.49328 0.234939 9.78539 -1.42846 0.013685 0.00148646 -0.0415218 1.0169 -50.3966 43.1768 1.0 0.0 0.0 0.0 30.3125 9.6937 -1.57902 -0.215687 9.6167 -1.6163 0.280488 -0.00657665 -0.00463778 3.6825E-4 -59.8479 -38.8919 -58.5253 1.0 0.0 0.0 0.0
Time taken: 0.191 seconds, Fetched: 2 row(s)
Create a HIVE managed ORC table to convert into orc format
hive> CREATE TABLE test1.pamap2_orc
(time DOUBLE, activity DOUBLE, heart_rate DOUBLE,hand_temp DOUBLE,hand_acc16g_1 DOUBLE, hand_acc16g_2 DOUBLE, hand_acc16g_3 DOUBLE, hand_acc6g_1 DOUBLE, hand_acc6g_2 DOUBLE, hand_acc6g_3 DOUBLE, hand_gyro_1 DOUBLE,hand_gyro_2 DOUBLE,hand_gyro_3 DOUBLE, hand_mag_1 DOUBLE, hand_mag_2 DOUBLE, hand_mag_3 DOUBLE, hand_orient_1 DOUBLE, hand_orient_2 DOUBLE, hand_orient_3 DOUBLE, hand_orient_4 DOUBLE,
chest_temp DOUBLE,chest_acc16g_1 DOUBLE, chest_acc16g_2 DOUBLE, chest_acc16g_3 DOUBLE, chest_acc6g_1 DOUBLE, chest_acc6g_2 DOUBLE, chest_acc6g_3 DOUBLE, chest_gyro_1 DOUBLE,chest_gyro_2 DOUBLE,chest_gyro_3 DOUBLE, chest_mag_1 DOUBLE, chest_mag_2 DOUBLE, chest_mag_3 DOUBLE, chest_orient_1 DOUBLE, chest_orient_2 DOUBLE, chest_orient_3 DOUBLE, chest_orient_4 DOUBLE,
ankle_temp DOUBLE,ankle_acc16g_1 DOUBLE, ankle_acc16g_2 DOUBLE, ankle_acc16g_3 DOUBLE, ankle_acc6g_1 DOUBLE, ankle_acc6g_2 DOUBLE, ankle_acc6g_3 DOUBLE, ankle_gyro_1 DOUBLE,ankle_gyro_2 DOUBLE,ankle_gyro_3 DOUBLE, ankle_mag_1 DOUBLE, ankle_mag_2 DOUBLE, ankle_mag_3 DOUBLE, ankle_orient_1 DOUBLE, ankle_orient_2 DOUBLE, ankle_orient_3 DOUBLE, ankle_orient_4 DOUBLE )
STORED AS ORC;
OK
As we have already loaded temporary table pampa2_table, it’s time to load the data from it to actual ORC table pampa2_orc.
Insert the data from the external table into the Hive-managed table.
hive> INSERT INTO TABLE test1.pamap2_orc SELECT * FROM test1.pamap2_table;
Query ID = hadoop_20180314092522_71507bf7-1c30-4376-87cd-b38129a7f6a9
Total jobs = 1
Launching Job 1 out of 1
Number of reduce tasks is set to 0 since there's no reduce operator
Starting Job = job_1552283641705_0251, Tracking URL = http://test-ser-vm01:8088/proxy/application_1552283641705_0251/
Kill Command = /mnt/data/hadoop/hadoop/bin/hadoop job -kill job_1552283641705_0251
Hadoop job information for Stage-1: number of mappers: 9; number of reducers: 0
2018-03-14 09:25:56,207 Stage-1 map = 0%, reduce = 0%
2018-03-14 09:26:05,033 Stage-1 map = 11%, reduce = 0%, Cumulative CPU 6.13 sec
2018-03-14 09:26:13,402 Stage-1 map = 22%, reduce = 0%, Cumulative CPU 11.18 sec
2018-03-14 09:26:14,445 Stage-1 map = 33%, reduce = 0%, Cumulative CPU 19.27 sec
2018-03-14 09:26:22,804 Stage-1 map = 66%, reduce = 0%, Cumulative CPU 84.16 sec
2018-03-14 09:26:23,846 Stage-1 map = 80%, reduce = 0%, Cumulative CPU 100.77 sec
2018-03-14 09:26:24,889 Stage-1 map = 88%, reduce = 0%, Cumulative CPU 119.36 sec
2018-03-14 09:26:30,099 Stage-1 map = 90%, reduce = 0%, Cumulative CPU 132.35 sec
2018-03-14 09:26:32,183 Stage-1 map = 96%, reduce = 0%, Cumulative CPU 135.55 sec
2018-03-14 09:26:38,455 Stage-1 map = 100%, reduce = 0%, Cumulative CPU 143.93 sec
MapReduce Total cumulative CPU time: 2 minutes 23 seconds 930 msec
Ended Job = job_1552283641705_0251
Stage-4 is selected by condition resolver.
Stage-3 is filtered out by condition resolver.
Stage-5 is filtered out by condition resolver.
Moving data to directory hdfs://test-ser-vm01:9000/user/hive/warehouse/test1.db/pamap2_orc/.hive-staging_hive_2018-03-14_09-25-22_801_2601980388252549428-1/-ext-10000
Loading data to table test1.pamap2_orc
MapReduce Jobs Launched:
Stage-Stage-1: Map: 9 Cumulative CPU: 143.93 sec HDFS Read: 1302077635 HDFS Write: 772976727 SUCCESS
Total MapReduce CPU Time Spent: 2 minutes 23 seconds 930 msec
OK
Time taken: 78.615 seconds
Verify the data import into ORC-formatted table:
hive> select * from test1.pamap2_orc limit 2;
OK
5.64 0.0 NaN 33.0 2.79143 7.55389 -7.06374 2.87553 7.88823 -6.76139 1.0164 -0.28941 1.38207 -11.6508 -3.73683 31.17841.0 0.0 0.0 0.0 36.125 1.94739 9.59644 -3.12873 1.81868 9.49711 -2.91989 0.124025 0.112482 -0.0449469 -20.2905 -32.0492 8.67906 1.0 0.0 0.0 0.0 33.8125 9.84408 -0.808951 -1.64674 9.73055 -0.846832 -1.29665 -0.027148 -0.0311901 -0.0408973 -47.7695 -2.58701 59.8481 -0.0128709 0.747947 -0.0798406 0.658813
5.65 0.0 NaN 33.0 2.86086 7.43814 -7.21626 2.84248 7.63164 -6.8514 1.08269 -0.393965 1.60935 -11.6575 -3.18648 30.7215 1.0 0.0 0.0 0.0 36.125 1.7512 9.6334 -3.32601 1.74445 9.69355 -2.96421 0.132679 0.0608292 -0.0441676 -20.6409 -31.69898.30648 1.0 0.0 0.0 0.0 33.8125 9.83968 -0.807666 -1.80115 9.73049 -0.816601 -1.31189 0.0128035 -0.0363842 -0.0148455 -47.7624 -2.81438 60.3407 0.0140248 -0.74841 0.0790426 -0.65836
Time taken: 0.158 seconds, Fetched: 2 row(s)
Successfuly converted into ORC format
hive> describe formatted test1.pamap2_orc;
OK
# col_name data_type comment
time double
activity double
heart_rate double
hand_temp double
hand_acc16g_1 double
hand_acc16g_2 double
. .
. .
. .
ankle_orient_3 double
ankle_orient_4 double
# Detailed Table Information
Database: test1
Owner: hadoop
CreateTime: Thu Mar 14 09:24:11 UTC 2018
LastAccessTime: UNKNOWN
Retention: 0
Location: hdfs://test-ser-vm01:9000/user/hive/warehouse/test1.db/pamap2_orc
Table Type: MANAGED_TABLE
Table Parameters:
COLUMN_STATS_ACCURATE {\"BASIC_STATS\":\"true\"}
numFiles 9
numRows 2872533
rawDataSize 1240934256
totalSize 772975987
transient_lastDdlTime 1552555601
# Storage Information
SerDe Library: org.apache.hadoop.hive.ql.io.orc.OrcSerde
InputFormat: org.apache.hadoop.hive.ql.io.orc.OrcInputFormat
OutputFormat: org.apache.hadoop.hive.ql.io.orc.OrcOutputFormat
Compressed: No
Num Buckets: -1
Bucket Columns: []
Sort Columns: []
Storage Desc Params:
serialization.format 1
Time taken: 0.604 seconds, Fetched: 83 row(s)
[hadoop@test-ser-vm01 Protocol]$ hadoop fs -ls /user/hive/warehouse/test1.db/pamap2_orc
Found 9 items
-rwxr-xr-x 3 hadoop supergroup 202673523 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000000_0
-rwxr-xr-x 3 hadoop supergroup 153111161 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000001_0
-rwxr-xr-x 3 hadoop supergroup 104919944 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000002_0
-rwxr-xr-x 3 hadoop supergroup 101485801 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000003_0
-rwxr-xr-x 3 hadoop supergroup 86684110 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000004_0
-rwxr-xr-x 3 hadoop supergroup 81040336 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000005_0
-rwxr-xr-x 3 hadoop supergroup 27038160 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000006_0
-rwxr-xr-x 3 hadoop supergroup 11794464 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000007_0
-rwxr-xr-x 3 hadoop supergroup 4228488 2018-03-14 09:26 /user/hive/warehouse/test1.db/pamap2_orc/000008_0
Pyspark for data processing and Machine Learning
Utilize SparkSQL and ml library to perform data pre-processing steps and machine learning to perform classification,
Importing necessary modules
from pyspark.sql import SparkSession
from pyspark import SparkConf, SparkContext
import pyspark.sql.functions as fn
from pyspark.ml.classification import RandomForestClassifier
from pyspark.ml.clustering import KMeans,BisectingKMeans
from pyspark.ml.feature import StringIndexer,OneHotEncoder,VectorAssembler,PCA as sparkpca,MinMaxScaler as sparkMinMaxScaler,Normalizer,VectorIndexer
from pyspark.ml import Pipeline as SparkPipeline,PipelineModel
import pandas as pd
from matplotlib import pyplot as plt
from pyspark.sql.types import DateType
from pyspark.ml.evaluation import RegressionEvaluator,MulticlassClassificationEvaluator,BinaryClassificationEvaluator
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
from pyspark.sql.types import *
from datetime import datetime
import numpy as np
%matplotlib inline
from pyspark.sql.types import *
from pyspark.sql.functions import array, explode, lit
import warnings
from operator import add
import re
from functools import reduce
warnings.filterwarnings('ignore')
Creating SparkSession object and reading orc input file
spark = SparkSession.builder.getOrCreate()
inputdataraw = spark.read.orc('/user/hive/warehouse/test1.db/pamap2_orc')
inputdataraw.cache()
inputdataraw.count()
2872533
inputdataraw.groupBy('activity').agg(fn.count('time')).show()
+--------+-----------+
|activity|count(time)|
+--------+-----------+
| 0.0| 929661|
| 7.0| 188107|
| 1.0| 192523|
| 4.0| 238761|
| 3.0| 189931|
| 2.0| 185188|
| 17.0| 238690|
| 13.0| 104944|
| 6.0| 164600|
| 24.0| 49360|
| 5.0| 98199|
| 16.0| 175353|
| 12.0| 117216|
+--------+-----------+
inputdataraw.printSchema()
root
|-- time: double (nullable = true)
|-- activity: double (nullable = true)
|-- heart_rate: double (nullable = true)
|-- hand_temp: double (nullable = true)
|-- hand_acc16g_1: double (nullable = true)
|-- hand_acc16g_2: double (nullable = true)
|-- hand_acc16g_3: double (nullable = true)
|-- hand_acc6g_1: double (nullable = true)
|-- hand_acc6g_2: double (nullable = true)
|-- hand_acc6g_3: double (nullable = true)
|-- hand_gyro_1: double (nullable = true)
|-- hand_gyro_2: double (nullable = true)
|-- hand_gyro_3: double (nullable = true)
|-- hand_mag_1: double (nullable = true)
|-- hand_mag_2: double (nullable = true)
|-- hand_mag_3: double (nullable = true)
|-- hand_orient_1: double (nullable = true)
|-- hand_orient_2: double (nullable = true)
|-- hand_orient_3: double (nullable = true)
|-- hand_orient_4: double (nullable = true)
|-- chest_temp: double (nullable = true)
|-- chest_acc16g_1: double (nullable = true)
|-- chest_acc16g_2: double (nullable = true)
|-- chest_acc16g_3: double (nullable = true)
|-- chest_acc6g_1: double (nullable = true)
|-- chest_acc6g_2: double (nullable = true)
|-- chest_acc6g_3: double (nullable = true)
|-- chest_gyro_1: double (nullable = true)
|-- chest_gyro_2: double (nullable = true)
|-- chest_gyro_3: double (nullable = true)
|-- chest_mag_1: double (nullable = true)
|-- chest_mag_2: double (nullable = true)
|-- chest_mag_3: double (nullable = true)
|-- chest_orient_1: double (nullable = true)
|-- chest_orient_2: double (nullable = true)
|-- chest_orient_3: double (nullable = true)
|-- chest_orient_4: double (nullable = true)
|-- ankle_temp: double (nullable = true)
|-- ankle_acc16g_1: double (nullable = true)
|-- ankle_acc16g_2: double (nullable = true)
|-- ankle_acc16g_3: double (nullable = true)
|-- ankle_acc6g_1: double (nullable = true)
|-- ankle_acc6g_2: double (nullable = true)
|-- ankle_acc6g_3: double (nullable = true)
|-- ankle_gyro_1: double (nullable = true)
|-- ankle_gyro_2: double (nullable = true)
|-- ankle_gyro_3: double (nullable = true)
|-- ankle_mag_1: double (nullable = true)
|-- ankle_mag_2: double (nullable = true)
|-- ankle_mag_3: double (nullable = true)
|-- ankle_orient_1: double (nullable = true)
|-- ankle_orient_2: double (nullable = true)
|-- ankle_orient_3: double (nullable = true)
|-- ankle_orient_4: double (nullable = true)
inputdataraw.describe().toPandas()
| summary | time | activity | heart_rate | hand_temp | hand_acc16g_1 | hand_acc16g_2 | hand_acc16g_3 | hand_acc6g_1 | hand_acc6g_2 | ... | ankle_gyro_1 | ankle_gyro_2 | ankle_gyro_3 | ankle_mag_1 | ankle_mag_2 | ankle_mag_3 | ankle_orient_1 | ankle_orient_2 | ankle_orient_3 | ankle_orient_4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | count | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | ... | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 | 2872533 |
| 1 | mean | 1834.3538678511266 | 5.4662425114002176 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | stddev | 1105.6889982931357 | 6.331333413102728 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | min | 5.64 | 0.0 | 57.0 | 24.75 | -145.367 | -104.301 | -101.452 | -61.4895 | -61.868 | ... | -23.995 | -18.1269 | -14.0196 | -172.865 | -137.908 | -109.289 | -0.253628 | -0.956876 | -0.876838 | -0.997281 |
| 4 | max | 4475.63 | 24.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
5 rows × 55 columns
inputdataraw.show(1)
+----+--------+----------+---------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+----------+----------+----------+-------------+-------------+-------------+-------------+----------+--------------+--------------+--------------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+--------------+--------------+--------------+--------------+----------+--------------+--------------+--------------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+--------------+--------------+--------------+--------------+
|time|activity|heart_rate|hand_temp|hand_acc16g_1|hand_acc16g_2|hand_acc16g_3|hand_acc6g_1|hand_acc6g_2|hand_acc6g_3|hand_gyro_1|hand_gyro_2|hand_gyro_3|hand_mag_1|hand_mag_2|hand_mag_3|hand_orient_1|hand_orient_2|hand_orient_3|hand_orient_4|chest_temp|chest_acc16g_1|chest_acc16g_2|chest_acc16g_3|chest_acc6g_1|chest_acc6g_2|chest_acc6g_3|chest_gyro_1|chest_gyro_2|chest_gyro_3|chest_mag_1|chest_mag_2|chest_mag_3|chest_orient_1|chest_orient_2|chest_orient_3|chest_orient_4|ankle_temp|ankle_acc16g_1|ankle_acc16g_2|ankle_acc16g_3|ankle_acc6g_1|ankle_acc6g_2|ankle_acc6g_3|ankle_gyro_1|ankle_gyro_2|ankle_gyro_3|ankle_mag_1|ankle_mag_2|ankle_mag_3|ankle_orient_1|ankle_orient_2|ankle_orient_3|ankle_orient_4|
+----+--------+----------+---------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+----------+----------+----------+-------------+-------------+-------------+-------------+----------+--------------+--------------+--------------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+--------------+--------------+--------------+--------------+----------+--------------+--------------+--------------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+--------------+--------------+--------------+--------------+
|5.64| 0.0| NaN| 33.0| 2.79143| 7.55389| -7.06374| 2.87553| 7.88823| -6.76139| 1.0164| -0.28941| 1.38207| -11.6508| -3.73683| 31.1784| 1.0| 0.0| 0.0| 0.0| 36.125| 1.94739| 9.59644| -3.12873| 1.81868| 9.49711| -2.91989| 0.124025| 0.112482| -0.0449469| -20.2905| -32.0492| 8.67906| 1.0| 0.0| 0.0| 0.0| 33.8125| 9.84408| -0.808951| -1.64674| 9.73055| -0.846832| -1.29665| -0.027148| -0.0311901| -0.0408973| -47.7695| -2.58701| 59.8481| -0.0128709| 0.747947| -0.0798406| 0.658813|
+----+--------+----------+---------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+----------+----------+----------+-------------+-------------+-------------+-------------+----------+--------------+--------------+--------------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+--------------+--------------+--------------+--------------+----------+--------------+--------------+--------------+-------------+-------------+-------------+------------+------------+------------+-----------+-----------+-----------+--------------+--------------+--------------+--------------+
only showing top 1 row
Referring to the activity id mappings we can infer that activity id = 0 should not be considered for the analysis as it indicates transient movement between different activities.Lets filter them out.
inputdataraw = inputdataraw.withColumn('new_time',fn.round(fn.col('time')%10))
inputdata = inputdataraw.filter(fn.col('activity')!=0)
inputdata = inputdata.withColumn('activity',fn.col('activity').cast('int'))
inputdata.select('activity').distinct().orderBy('activity').show()
+--------+
|activity|
+--------+
| 1|
| 2|
| 3|
| 4|
| 5|
| 6|
| 7|
| 12|
| 13|
| 16|
| 17|
| 24|
+--------+
We know that heart rate would be considered as an important measure to identify the nature of physical activity,lets group the data by activity and check mean within each group to identify any pattern and validate our hypothesis
heart_rate_agg = inputdata.filter(~fn.isnan('heart_rate')).groupBy('activity').agg(fn.mean('heart_rate').alias('avg_heart_rate'))
heart_rate_agg.sort(fn.desc('avg_heart_rate')).show()
+--------+------------------+
|activity| avg_heart_rate|
+--------+------------------+
| 24|161.98139122729287|
| 5|156.59581411049848|
| 12|129.52348491922683|
| 13|129.15417491921193|
| 6| 124.8799521403882|
| 7|123.82870101174555|
| 4|112.78986505095016|
| 16| 104.1980908410282|
| 17| 90.06959118052366|
| 3| 88.55763688760807|
| 2| 80.01258195995038|
| 1| 75.53568181818181|
+--------+------------------+
Indeed low intensity activities and high intensity activities have varying heart rates as expected.
Checking and treating for Null values
from functools import reduce
null_counts =reduce(lambda x,y:x.union(y),(inputdata.agg(fn.count(fn.when(fn.isnan(c), c)).alias('null_count')).select(fn.lit(c).alias('col_name'),'null_count') for c in inputdata.columns))
Missing sensory data due to wireless data dropping: missing values are indicated with NaN.It is a case of missingness completely at random.
null_counts.sort(fn.desc('null_count')).show(100)
+--------------+----------+
| col_name|null_count|
+--------------+----------+
| heart_rate| 1765464|
| hand_acc16g_2| 11124|
| hand_orient_4| 11124|
| hand_orient_1| 11124|
| hand_mag_1| 11124|
| hand_acc6g_3| 11124|
| hand_acc6g_2| 11124|
| hand_temp| 11124|
| hand_gyro_2| 11124|
| hand_orient_3| 11124|
| hand_acc16g_1| 11124|
| hand_acc16g_3| 11124|
| hand_orient_2| 11124|
| hand_gyro_3| 11124|
| hand_acc6g_1| 11124|
| hand_gyro_1| 11124|
| hand_mag_3| 11124|
| hand_mag_2| 11124|
|ankle_orient_1| 8507|
| ankle_acc6g_2| 8507|
|ankle_orient_4| 8507|
| ankle_gyro_2| 8507|
|ankle_acc16g_1| 8507|
| ankle_mag_1| 8507|
|ankle_orient_3| 8507|
|ankle_acc16g_2| 8507|
|ankle_acc16g_3| 8507|
| ankle_gyro_3| 8507|
|ankle_orient_2| 8507|
| ankle_acc6g_3| 8507|
| ankle_mag_3| 8507|
| ankle_mag_2| 8507|
| ankle_acc6g_1| 8507|
| ankle_temp| 8507|
| ankle_gyro_1| 8507|
| chest_gyro_1| 2420|
| chest_acc6g_2| 2420|
| chest_mag_1| 2420|
|chest_orient_2| 2420|
|chest_acc16g_3| 2420|
| chest_mag_2| 2420|
| chest_acc6g_1| 2420|
| chest_gyro_2| 2420|
| chest_mag_3| 2420|
| chest_acc6g_3| 2420|
| chest_temp| 2420|
|chest_orient_3| 2420|
|chest_orient_4| 2420|
|chest_acc16g_1| 2420|
|chest_orient_1| 2420|
| chest_gyro_3| 2420|
|chest_acc16g_2| 2420|
| time| 0|
| activity| 0|
+--------------+----------+
As we know the sensory data is time framed we can forward-fill and backward-fill null values using SparkSQL Window function - partition by activity and order by time
for column in inputdata.columns:
inputdata = inputdata.withColumn(column,fn.when(fn.isnan(fn.col(column)),None).otherwise(fn.col(column)))
from pyspark.sql import Window
import sys
window_ffill = Window.partitionBy('activity').orderBy('time').rowsBetween(-sys.maxsize,0)
window_bfill = Window.partitionBy('activity').orderBy('time').rowsBetween(0,sys.maxsize)
for c in [x for x in inputdata.columns if x not in ['time','activity']]:
inputdata = inputdata.withColumn(c,fn.last(fn.col(c),ignorenulls=True).over(window_ffill))
for c in [x for x in inputdata.columns if x not in ['time','activity']]:
inputdata = inputdata.withColumn(c,fn.last(fn.col(c),ignorenulls=True).over(window_bfill))
null_counts_check =reduce(lambda x,y:x.union(y),(inputdata.agg(fn.count(fn.when(fn.isnull(c), c)).alias('null_count')).select(fn.lit(c).alias('col_name'),'null_count') for c in inputdata.columns))
null_counts_check.sort(fn.desc('null_count')).show()
+--------------+----------+
| col_name|null_count|
+--------------+----------+
|ankle_acc16g_3| 0|
| hand_temp| 0|
| ankle_acc6g_1| 0|
| hand_orient_2| 0|
|chest_acc16g_1| 0|
| ankle_gyro_1| 0|
| hand_gyro_3| 0|
| ankle_acc6g_3| 0|
| hand_orient_3| 0|
| ankle_mag_3| 0|
| activity| 0|
| chest_mag_2| 0|
|chest_orient_2| 0|
| ankle_acc6g_2| 0|
|ankle_acc16g_1| 0|
| chest_acc6g_3| 0|
| chest_gyro_1| 0|
| hand_acc16g_1| 0|
| chest_gyro_2| 0|
| chest_acc6g_2| 0|
+--------------+----------+
only showing top 20 rows
Feature engineering
For each body sensor measurement available lets compare each value and generate aggregated features such as mean,standard deviation,min,max,max-min etc.
##feature engineering
hand_cols = [col for col in inputdata.columns if re.search(r"hand",col)]
chest_cols = [col for col in inputdata.columns if re.search(r"chest",col)]
ankle_cols = [col for col in inputdata.columns if re.search(r"ankle",col)]
sensor_cols = ['temp',
'acc16g_1',
'acc16g_2',
'acc16g_3',
'acc6g_1',
'acc6g_2',
'acc6g_3',
'gyro_1',
'gyro_2',
'gyro_3',
'mag_1',
'mag_2',
'mag_3',
'orient_1',
'orient_2',
'orient_3',
'orient_4']
for col_nm,a,b,c in zip(sensor_cols,hand_cols,chest_cols,ankle_cols):
inputdata = inputdata.withColumn(col_nm+'all_sum',fn.col(a)+fn.col(b)+fn.col(c))
inputdata = inputdata.withColumn(col_nm+'all_max',fn.greatest(fn.col(a),fn.col(b),fn.col(c)))
inputdata = inputdata.withColumn(col_nm+'all_min',fn.least(fn.col(a),fn.col(b),fn.col(c)))
inputdata = inputdata.withColumn(col_nm+'all_max-min',fn.col(col_nm+'all_max')-fn.col(col_nm+'all_min'))
inputdata = inputdata.withColumn(col_nm+'all_mean',fn.col(col_nm+'all_sum')/3)
inputdata = inputdata.withColumn(col_nm+'all_std',fn.sqrt(reduce(add, ((fn.col(x) - fn.col(col_nm+'all_mean')) ** 2 for x in [a,b,c])) /2))
Preparing target variable
From the activity list we can group the activities based on the intensity of the physical activity carried out into minimal,low,medium,high intensity bins for classification purpose.
#Minimun: [lying,sitting,standing,computer work,car driving,folding laundry]
minimum_activity = [1,2,3,9,10,11,18]
#Low: [descending stairs, ironing, house cleaning]
low_activity = [13,17,19]
#medium: [walking,descending stairs,vacuum cleaning]
medium_activity = [4,12,16]
#high: [running,cycling,nordic walking,playing soccer,rope jumping]
high_activity = [5,6,7,20,24]
##target labels
inputdata = inputdata.withColumn('target',fn.when(fn.col('activity').isin(minimum_activity),'min'))
inputdata = inputdata.withColumn('target',fn.when(fn.col('activity').isin(low_activity),'low').otherwise(fn.col('target')))
inputdata = inputdata.withColumn('target',fn.when(fn.col('activity').isin(medium_activity),'medium').otherwise(fn.col('target')))
inputdata = inputdata.withColumn('target',fn.when(fn.col('activity').isin(high_activity),'high').otherwise(fn.col('target')))
inputdata.select(fn.col('target')).distinct().show()
+------+
|target|
+------+
| low|
| high|
|medium|
| min|
+------+
Perform label indexing for each target variable
inputdata = inputdata.withColumn('label',fn.when(fn.col('target')=='min',0))
inputdata = inputdata.withColumn('label',fn.when(fn.col('target')=='low',1).otherwise(fn.col('label')))
inputdata = inputdata.withColumn('label',fn.when(fn.col('target')=='medium',2).otherwise(fn.col('label')))
inputdata = inputdata.withColumn('label',fn.when(fn.col('target')=='high',3).otherwise(fn.col('label')))
Machine Learning using pyspark
Utlizing machine learning to detect the intensity of physical activity using the readings of wearable devices that can track body motions and basic physiological parameters such as heart rate,temperature,acceleration,gyro,orientation parameters of different parts of the body.
Now we have our data processing and transformation using SparkSQL completed,lets perform machine learning using Pyspark ml library
inputdata.cache()
inputdata.count()
1942872
inputdata.select('target','label').distinct().show()
+------+-----+
|target|label|
+------+-----+
| high| 3|
| min| 0|
| low| 1|
|medium| 2|
+------+-----+
Preparing train/test data and features/target
train,test = inputdata.randomSplit([0.8, 0.2])
features = ['heart_rate','hand_temp','hand_acc16g_1','hand_acc16g_2','hand_acc16g_3','hand_acc6g_1','hand_acc6g_2','hand_acc6g_3','hand_gyro_1',
'hand_gyro_2','hand_gyro_3','hand_mag_1','hand_mag_2','hand_mag_3','hand_orient_1','hand_orient_2','hand_orient_3','hand_orient_4',
'chest_temp','chest_acc16g_1','chest_acc16g_2','chest_acc16g_3','chest_acc6g_1','chest_acc6g_2','chest_acc6g_3','chest_gyro_1',
'chest_gyro_2','chest_gyro_3','chest_mag_1','chest_mag_2','chest_mag_3','chest_orient_1','chest_orient_2','chest_orient_3',
'chest_orient_4','ankle_temp','ankle_acc16g_1','ankle_acc16g_2','ankle_acc16g_3','ankle_acc6g_1','ankle_acc6g_2','ankle_acc6g_3',
'ankle_gyro_1','ankle_gyro_2','ankle_gyro_3','ankle_mag_1','ankle_mag_2','ankle_mag_3','ankle_orient_1','ankle_orient_2',
'ankle_orient_3','ankle_orient_4','tempall_sum','tempall_max','tempall_min','tempall_max-min','tempall_mean',
'tempall_std','acc16g_1all_sum','acc16g_1all_max','acc16g_1all_min','acc16g_1all_max-min','acc16g_1all_mean','acc16g_1all_std',
'acc16g_2all_sum','acc16g_2all_max','acc16g_2all_min','acc16g_2all_max-min','acc16g_2all_mean','acc16g_2all_std','acc16g_3all_sum',
'acc16g_3all_max','acc16g_3all_min','acc16g_3all_max-min','acc16g_3all_mean','acc16g_3all_std','acc6g_1all_sum','acc6g_1all_max',
'acc6g_1all_min','acc6g_1all_max-min','acc6g_1all_mean','acc6g_1all_std','acc6g_2all_sum','acc6g_2all_max','acc6g_2all_min',
'acc6g_2all_max-min','acc6g_2all_mean','acc6g_2all_std','acc6g_3all_sum','acc6g_3all_max','acc6g_3all_min','acc6g_3all_max-min',
'acc6g_3all_mean','acc6g_3all_std','gyro_1all_sum','gyro_1all_max','gyro_1all_min','gyro_1all_max-min','gyro_1all_mean','gyro_1all_std',
'gyro_2all_sum','gyro_2all_max','gyro_2all_min','gyro_2all_max-min','gyro_2all_mean','gyro_2all_std','gyro_3all_sum','gyro_3all_max',
'gyro_3all_min','gyro_3all_max-min','gyro_3all_mean','gyro_3all_std','mag_1all_sum','mag_1all_max','mag_1all_min','mag_1all_max-min',
'mag_1all_mean','mag_1all_std','mag_2all_sum','mag_2all_max','mag_2all_min','mag_2all_max-min','mag_2all_mean','mag_2all_std',
'mag_3all_sum','mag_3all_max','mag_3all_min','mag_3all_max-min','mag_3all_mean','mag_3all_std','orient_1all_sum','orient_1all_max',
'orient_1all_min','orient_1all_max-min','orient_1all_mean','orient_1all_std','orient_2all_sum','orient_2all_max','orient_2all_min',
'orient_2all_max-min','orient_2all_mean','orient_2all_std','orient_3all_sum','orient_3all_max','orient_3all_min','orient_3all_max-min',
'orient_3all_mean','orient_3all_std','orient_4all_sum','orient_4all_max','orient_4all_min','orient_4all_max-min','orient_4all_mean',
'orient_4all_std']
target = 'label'
Random forest with Cross-validation using Pyspark
def spark_rf_classifier(train, test,features,label):
mlassembler = VectorAssembler(inputCols=features,outputCol="features")
rf = RandomForestClassifier(labelCol = label, featuresCol="features")
rfpipeline = SparkPipeline(stages=[mlassembler,rf])
rfparamGrid = ParamGridBuilder().addGrid(rf.maxDepth, [5,10,20]).addGrid(rf.numTrees, [100,250,500]).build()
rfevaluator = MulticlassClassificationEvaluator(labelCol =label,predictionCol = "prediction",metricName = "accuracy")
rfcrossval = CrossValidator(estimator=rfpipeline,
estimatorParamMaps=rfparamGrid,
evaluator=rfevaluator,
numFolds=3,parallelism=5)
rfmodel = rfcrossval.fit(train)
rfpredict = rfmodel.transform(test)
rf_accuracy = rfevaluator.evaluate(rfpredict,{rfevaluator.metricName: "accuracy"})
rf_precision = rfevaluator.evaluate(rfpredict,{rfevaluator.metricName: "weightedPrecision"})
rf_recall = rfevaluator.evaluate(rfpredict,{rfevaluator.metricName: "weightedRecall"})
rf_f1 = rfevaluator.evaluate(rfpredict,{rfevaluator.metricName: "f1"})
return ("Random Forest Model",rfmodel,rfpredict,rf_accuracy,rf_precision,rf_recall,rf_f1,rfevaluator,rfcrossval)
result = spark_rf_classifier(train, test,features,target)
print(result[1])
best_pipeline = result[1].bestModel
print(best_pipeline)
best_pipeline.stages
CrossValidatorModel_4b37928f1bbcf19ad86d
PipelineModel_474599db674a829f00e6
[VectorAssembler_409d85531cdc48ea7dd4,
RandomForestClassificationModel (uid=RandomForestClassifier_410bbd81e41f7e012fe4) with 100 trees]
From the cross-validation mechanism we have our best Random forest model,lets check the best paramters of the model
best_rf_model = best_pipeline.stages[1]
best_rf_model.getNumTrees
100
rf = RandomForestClassifier(labelCol = 'label', featuresCol="features")
paramGrid = ParamGridBuilder().addGrid(rf.maxDepth, [5,10,20]).addGrid(rf.numTrees, [100,250,500]).build()
java_model = best_rf_model._java_obj
{param.name: java_model.getOrDefault(java_model.getParam(param.name))
for param in paramGrid[0]}
{'maxDepth': 20, 'numTrees': 100}
We have performed hyperparameter tuning for the random forest model and indentified the hyperparameter with best validation metrics.
Training model and saving for prediction
mlassembler = VectorAssembler(inputCols=features,outputCol="features")
final_rf = RandomForestClassifier(maxDepth=20,numTrees=100,labelCol = 'label', featuresCol="features")
final_rfpipeline = SparkPipeline(stages=[mlassembler,final_rf])
final_rf_model = final_rfpipeline.fit(train)
final_rf_model.write().overwrite().save('/mnt/data/scripts/notebooks/ashwin/models/final_rf_model_trained')
model = PipelineModel.load('/mnt/data/scripts/notebooks/ashwin/models/final_rf_model_trained')
preds = model.transform(test)
preds.groupBy('label','prediction').count().show()
+-----+----------+------+
|label|prediction| count|
+-----+----------+------+
| 0| 0.0|113181|
| 1| 2.0| 250|
| 2| 3.0| 1|
| 3| 2.0| 44|
| 3| 3.0| 99606|
| 2| 2.0|106191|
| 1| 1.0| 68570|
+-----+----------+------+
As we can see most of the actual label and predictions are accurate and only a few are misclassified.This is a very good performing model for this classification purpose.
Extracting Feature importances
feat_imp = model.stages[-1].featureImportances
def ExtractFeatureImp(featureImp, dataset, featuresCol):
list_extract = []
for i in dataset.schema[featuresCol].metadata["ml_attr"]["attrs"]:
list_extract = list_extract + dataset.schema[featuresCol].metadata["ml_attr"]["attrs"][i]
varlist = pd.DataFrame(list_extract)
varlist['score'] = varlist['idx'].apply(lambda x: featureImp[x])
return(varlist.sort_values('score', ascending = False))
feature_importances = ExtractFeatureImp(feat_imp,predictions,"features")
feat_imp_df = pd.DataFrame(feature_importances).sort_values(by=['score'],ascending=False)
feat_imp_df.head()
| idx | name | score | |
|---|---|---|---|
| 0 | 0 | heart_rate | 0.155386 |
| 52 | 52 | tempall_sum | 0.039391 |
| 1 | 1 | hand_temp | 0.036727 |
| 54 | 54 | tempall_min | 0.035904 |
| 18 | 18 | chest_temp | 0.034430 |
feat_imp_df.drop(columns=['idx'],inplace=True)
feat_imp_df.set_index('name',inplace=True)
feat_imp_df.head(20).plot(kind='barh',figsize=(16,8))
plt.title('Feature Importances')
Text(0.5,1,'Feature Importances')
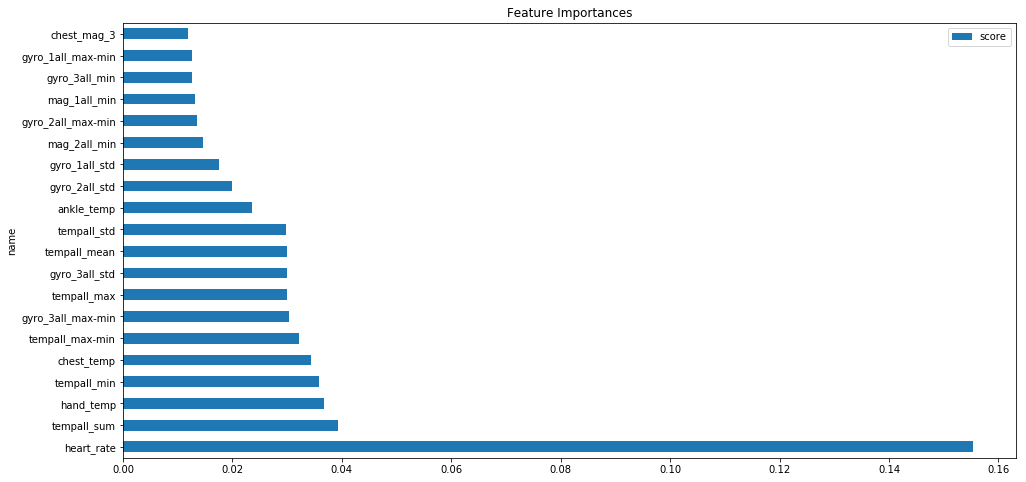
As we can see from the feature importances extracted heart rate and temperature related features play an important role in classification trees.Overall we can see the engineered features have good feature importances in our model compared to the original features.
eval_rf = BinaryClassificationEvaluator(rawPredictionCol='prediction', labelCol='label', metricName='areaUnderROC')
eval_rf.evaluate(preds)
1.0